Mass Spectrometry and Analytical Pharmacology Shared Resource (MSAPSR)
Proteomics Instruments and Services
A core mission of the MSAP is to accelerate discovery by giving investigators access to cutting edge technologies in mass spectrometry-based proteomics. By using advanced mass spectrometry and well-established proteomics technologies, we are devoted to help researchers to answer questions in basic, translational and clinical biomedical research. In addition, we host the Hyperion Imaging Mass Cytometry system which couples CyTOF technology with imaging for simultaneous detection of up to 40 protein markers in tissue sections.
Committed to providing researchers with high-quality, cost-effective services and support, we offer free consultation to each specific project, small- or large-scale, to ensure that expectations are properly managed and fulfilled. To keep scientists in our community competitive, new technologies will be incorporated into the facility as soon as possible to meet their needs. The facility is open to all investigators at Georgetown University and the LCCC Research Consortium, as well as outside GU.
The high resolution, sensitivity, and accuracy of our mass spectrometers allow accurate qualitative and quantitative measurements. The most common experiments include:
- Hypothesis-driven discovery proteomic studies of biological systems for the identification of peptides, proteins and post-translational modifications as well as their quantification by incorporating various strategies (e.g., iTRAQ-based stable isotopic labeling and label-free data independent quantification (DIA)
- Targeted proteomics by mainly using multiple reaction monitoring (MRM) and parallel reaction monitoring (PRM), e.g., for biomarker discovery and validation.
Proteomics Services offered by this shared resource include:
- Advice and guidance on the use and applications of methodologies and on experimental design
- Support for the writing of properly powered grant proposals and scientific manuscripts
- Research and development to fit users’ specific needs
- Training on the safe and proper use of instrumentation
- Mentoring of technicians and students involved in projects
Proteomics Technologies offered by this shared resource:
We provide a diverse range of services to characterize peptides/proteins from a variety of samples (including immunoprecipitates, whole cell lysates, tissues, biofluids, etc.), including but not limited to the following:
- Molecular weight determination of protein/peptide
- Protein identification from gel bands
- Protein identification from complex mixtures
- Characterization of post-translational modifications (PTMs): phosphorylation, acetylation, O-GlcNAcylation, etc.
- Protein quantification: Isobaric tagging for relative and absolute quantitation (iTRAQ) and label-free quantification (e.g., DIA-SWATH)
- Targeted quantification by Multiple Reaction Monitoring (MRM) and Parallel Reaction Monitoring (PRM)
- Others: 1-D gel electrophoresis, 2-D gel electrophoresis, MesoScale ECL ELISA
Proteomics & Hyperion Imaging Instrumentation
Orbitrap Lumos Tribrid mass spectrometer coupled with Ultimate 3000 HPLC (Thermo Fisher Scientific) or NanoUPLC Acquity (Waters): Used mainly for discovery proteomics/peptidomics

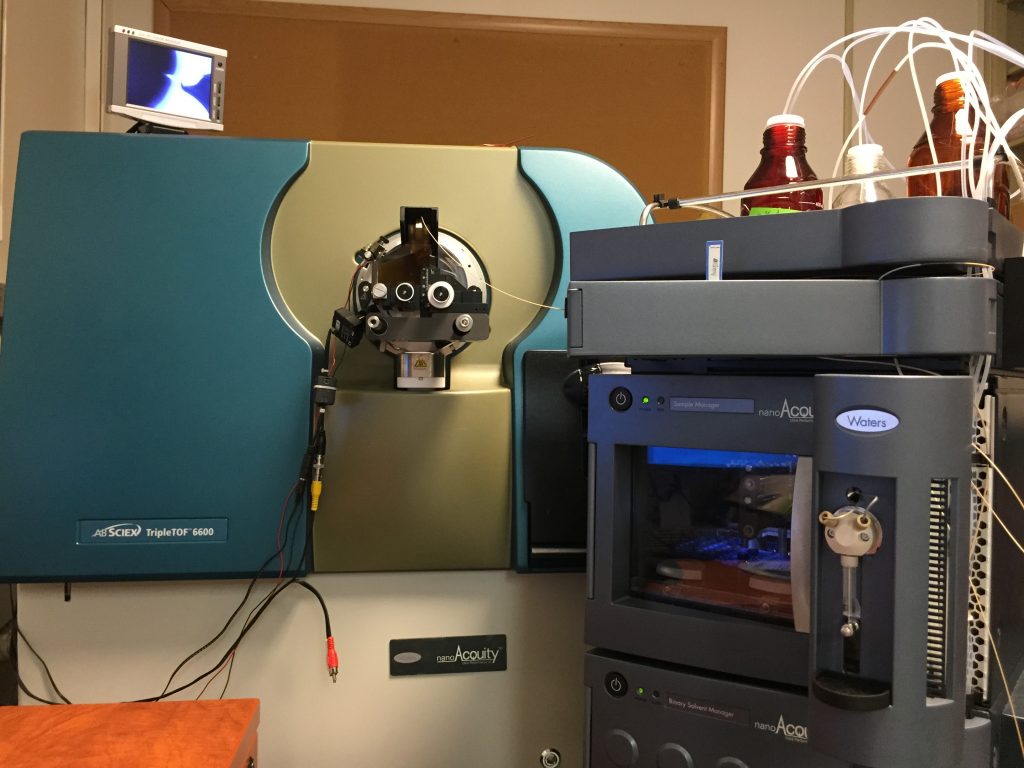
Triple TOF 6600 (Sciex) coupled with NanoUPLC Acquity (Waters): Used mainly for discovery proteomics/peptidomics
Q-TRAP 6500 (Sciex) coupled with NanoUPLC Acquity (Waters): Used for targeted proteomics/peptidomics

Hyperion Imaging Mass Cytometry (Fluidim): coupling CyTOF technology with imaging to provide simultaneous detection of up to 40 protein markers in tissue sections.

Classic Acquity UPLC (Waters): Used mainly for offline sample preparation

Barocycler NEP2320: Used for pressure-assisted sample

Meso Scale Discovery (MSD) Sector Imager: Used for immunoassays