Genomics and Epigenomics Shared Resource
Services and Instruments
Services offered by the GESR include:
- Data Acquisition: The GESR offers data acquisition services using a variety of instruments. Please complete the Service Request Form. We will contact you for more details to put together a quote for you. If you have already received a quote and would like to proceed with data acquisition, please complete the Sample Submission Form.
- Bioinformatics Support: The GESR provides bioinformatics support such as including analysis of data acquired by next generation sequencing (NGS), microarray, and qRT-PCR. Please complete the Bioinformatics Support Request Form. Furthermore, the GESR provides access to software tools such as Ingenuity Pathway Analysis (IPA) tool, Partek Genomics Suite, and Partek Flow. Please complete the Software Access Request Form.
- Access to User-Operated Instruments: The GESR houses several user-operated instruments. Please complete the user-operated instrument access request form. To schedule time to access to PCR machines, click here.
- Other support:
- Advice and guidance on the use and applications of methodologies and on experimental design
- Support for the writing of properly powered grant proposals and scientific manuscripts
- Research and development to fit users’ specific needs
- Training on the safe and proper use of user-operated instruments (e.g., RT-PCRs, plate readers, spectrophotometers, and densitometers)
- Mentoring of technicians and students involved in projects
Technologies available include:
Next-generation sequencing
Illumina NextSeq 550: The NextSeq 550 is one of the new sequencers from Illumina that use the 2-channel sequencing technology. With tunable output and high data quality, NextSeq provides the flexible power needed for whole-genome, transcriptome, and targeted resequencing plus the ability to scan microarrays including the Infinium MethylationEPIC BeadChip and other select BeadChips. The NextSeq 550 runs on 400M (high output) and 130M (mid output) flow cells with a maximum output of 120Gb.
Illumina MiSeq: The MiSeq is the only Illumina sequencer that allows 600 cycle reading length, which is critical for applications such as metagenomics that requires long reads. MiSeq focuses on applications such as targeted resequencing, metagenomics, small genome sequencing, targeted gene expression profiling. MiSeq runs on 25M (v3) and 15M (v2) flow cells with a maximum output of 15Gb.
Covaris M220 Focused-ultrasonicator: The Covaris M220 is used for DNA shearing for next-generation sequencing applications requiring fragment sizes between 150bp and 5kb.
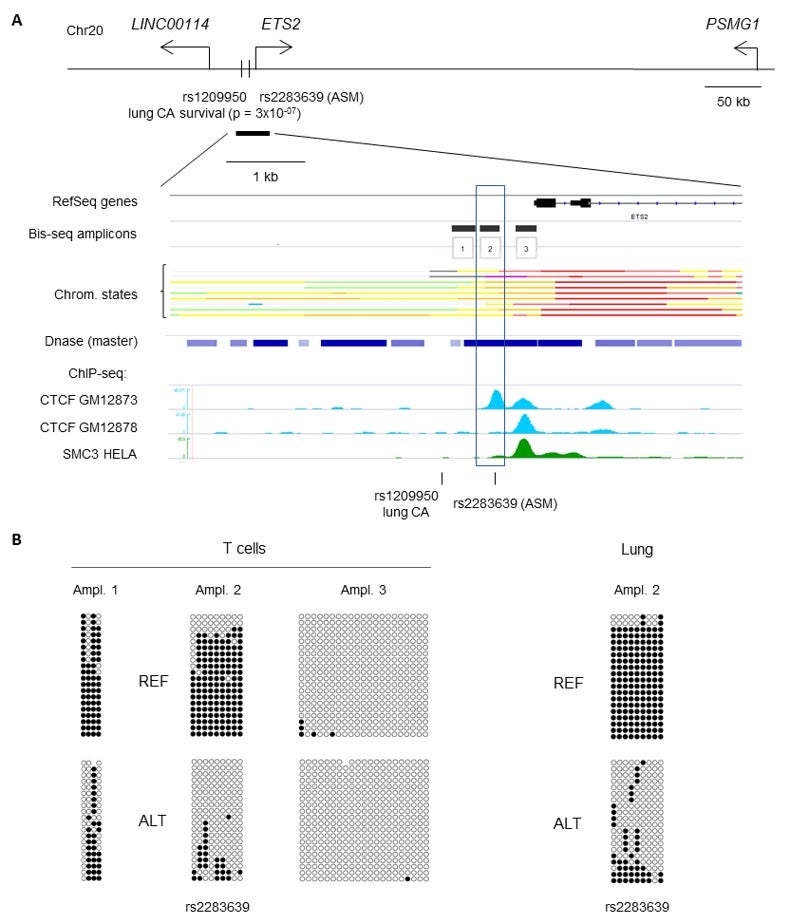
Nextgen Methyl-seq (bisulfite sequencing): The Fluidigm Juno System uses patented microfluidics technology to perform library preparation for targeted DNA sequencing on the Illumina system using an integrated fluidic circuit (IFC). The dynamic arrays have an integrated network of channels, chambers, and valves that automatically combine the reactions, conserving time reagent and sample amount. Using the Juno system, dozens to hundreds of samples can be processed for targeted Nextgen sequencing of hundreds of amplicons, for SNP and mutation detection, and/or targeted Nextgen Methyl-seq (bisulfite sequencing) for measuring and mapping net and allele-specific CpG methylation. An example of data generated by the Juno MiSEQ pipeline is in Figure 1.
Single-cell sequencing
10X Genomics Chromium Controller: The 10X Genomics Chromium Controller platform allows scientists to analyze genome or transcriptomes on a cell-by-cell basis through the use of microfluidic partitioning to capture single cells and prepare barcoded, NGS libraries. Furthermore, the addition of the Feature Barcoding technology allows scientists to simultaneously measure gene expression profiling and cell surface protein expression in the same cell. The Visium Spatial Gene Expression from 10x Genomics allows for high-resolution gene expression profiling for intact tissue sections. The overlay of the gene expression data with the H&E image for the same sample enables scientists to reveal information about the relationship of cellular function, phenotype, and location in tissue microenvironments.
Digital and Spatial Gene Expression Profiling
NanoString GeoMX Digital Spatial Profiler: GeoMx Protein and RNA assays enable spatially resolved, digital readout of up to 96 proteins or RNA targets in a multiplexed assay on a single FFPE or fresh frozen slide. The assay relies upon antibody or RNA probes coupled to photocleavable oligonucleotide tags. After hybridization of probes to slide-mounted formalin fixed paraffin-embedded (FFPE) tissue sections, the oligonucleotide tags are released from discrete regions of the tissue via UV exposure. Released tags are quantitated in a standard nCounter assay, and counts are mapped back to tissue location, yielding a spatially-resolved digital profile of analyte abundance.
Three microarray platforms (Affymetrix, Agilent, and Illumina) are used for mRNA/miRNA expression profiling, SNP genotyping, CNV analysis and DNA methylation analysis.
Affymetrix Microarray System: The Affymetrix system includes GeneChip scanner 3000 7G-Plus that allows scanning high-density microarrays.
Agilent Microarray System: The Agilent system is a high-resolution scanner that scans Agilent SurePrint G3 microarrays.
Illumina iScan: The Illumina system offers a cost-effective alternative to Affymetrix and Agilent microarray systems.
Real-Time PCR
QuantStudio Real-Time PCR (RT-PCR) with OpenArray block and Accufill System is used for mRNA/miRNA profiling and SNP genotyping with TaqMan probes.
StepOnePlus Real-Time PCR (RT-PCR) is used for mRNA/miRNA profiling and SNP genotyping with TaqMan probes.
Sample Quality Assessment and Assay Setup
Agilent Bioanalyzer 2100: Agilent Bioanalyzer 2100 (lab-on-a-chip) is used to perform DNA and RNA quality control tests prior to analysis of samples by microarray, NGS, NanoString and RT-PCR. The instrument provides a microfluidics-based platform for sizing, quantification and quality control of RNA, DNA, proteins and cells.
DNA plating and assay preparation using the Beckman Multimek NXP Robotic Liquid Handling System: The Biomek NXp is capable of performing a wide range of tasks, including assay preparation by transferring samples and reagents from tubes to 96- or 384-well plates (e.g., DNA plating and assay preparation for SNP genotyping).
The Mid-Atlantic Shared Resources Consortium provides access to additional instruments:
- Illumina HiSeq 4000: production-scale sequencing
- Pacific Biosciences RS II: Single Molecule, Real-Time (SMRT) Sequencing technology delivers highly accurate long reads.